-
Notifications
You must be signed in to change notification settings - Fork 18
MRI_Heights_of_Surfaces_Tools
The tools help to compare the height in the z-dimension of the signals in two different channels (red and blue). It calculates the fraction of the volume of the red signal that lies above the blue signal and the fraction of the red signal that lies below the blue signal. Only places where both signals are present (heights of red and blue bigger than zero) are taken into account. You can find a synthetic example image here: sdata03.tif.
The source code in git-hub can be found here.
To install the tools, drag the link heights_of_surfaces_tools.ijm to the FIJI launcher window, save it under macros/toolsets in the FIJI installation and restart FIJI.
Select the "heights of surfaces tools" toolset from the >> button of the ImageJ launcher.
- the first button (the one with an image) open this help page
- the b-button runs the background correction on the input image
- the c-button creates a 2D-surface image in which the pixel values correspond to the max. z-position at a point after the image has been thresholded
- the p-button will display a line-profile plot, containing the red and blue channels, on the surface image.
- the v-button will calculate the volume of the red signal above the blue signal and the volume of the gap between the red signal and the blue signal for places where the red signal is below the blue signal
- the last button (with the gear-icon) will run the batch processing on a folder containing the input images
Options are available for the background correction, the creation of the surface image and the batch-processing. The option dialogs can be opened by a right-click on the b, c and gear-buttons.
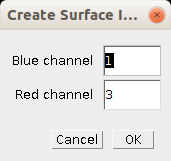
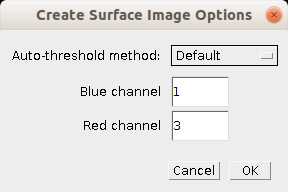
- Blue channel: The index of the blue channel in the image
- Red channel: The index of the red channel in the image
- Auto-threshold method: The method used to automatically determine the threshold for each channel
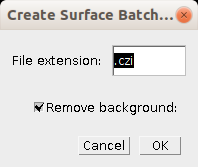
- File extension: The file extension of the input image files
- Remove background: If selected the background correction will be used in the batch processing
The method tries to find background regions and calculates the mean-background intensity per z-slice. The mean value of each z-slice is subtracted from the slice.
The image is thresholded and a new 2d-image is created, in which each pixel value represents the maximum z-position of the pixel for which the thresholded intensity is not zero.
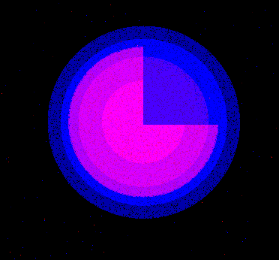
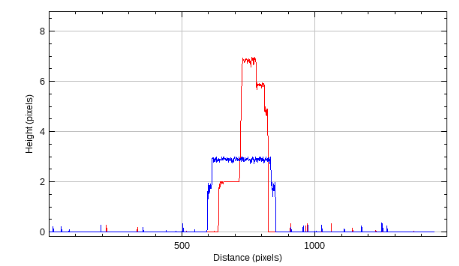
A line is drawn on the image and a line plot is displayed. The the x-axis represents the distance from the start of the line and the y-axis the maximum height of the signal. The result for both channels is displayed in the same plot. If the image already has a line selection when the command is run, the already existing selection is used, otherwise a default selection from the upper-right corner to the lower left corner is used.
The volume of the red channel above the blue channel and the volume of the blue channel above the red channel (which is in the same time the volume of the gap between the red channel and the blue channel when the red signal is lower than the blue one) are calculated and reported as fractions of the total volume and as absolute values. Only positions for which both signals have a height bigger than zero are taken into account.
When you run the batch-processing a first dialog will ask for the path to the folder containing the input images. All images that have the right file extension in the subtree of the input folder will be processed. A second dialog will ask for an output folder. The line plot, a control image of the 2D-surface image with the region that has been taken into account and the final results-table, containg the measurements of all images will be saved into this folder.



 Volker Bäcker
Volker Bäcker